At a glance

Measuring Transmission with Rt
The basic reproductive number, R0 (pronounced R-naught), is defined as the expected number of new infections caused by each infected person in a fully susceptible population and in the absence of interventions. R0 is an important theoretical concept in epidemiology, but in the real world, a fully susceptible population rarely exists.
The time-varying effective reproductive number, known as Rt, is defined as the average number of new infections caused by each currently infectious person—reflecting current population susceptibility, interventions, and variant transmissibility at the time it is measured. It is a data-driven metric, unlike R0. When Rt is above one, infections are increasing; when Rt is below one, infections are decreasing.
What Rt can and cannot tell us:
What Rt can tell us: Rt can tell us whether infections are increasing, decreasing, or remaining stable, and is an additional tool to help public health practitioners prepare and respond.
What Rt cannot tell us: Rt cannot tell us about the underlying burden of disease, just the trend of transmission. An Rt< 1 does not mean that transmission is low, just that infections are decreasing. It is useful to look at respiratory disease activity in conjunction with Rt.
What is Rt?
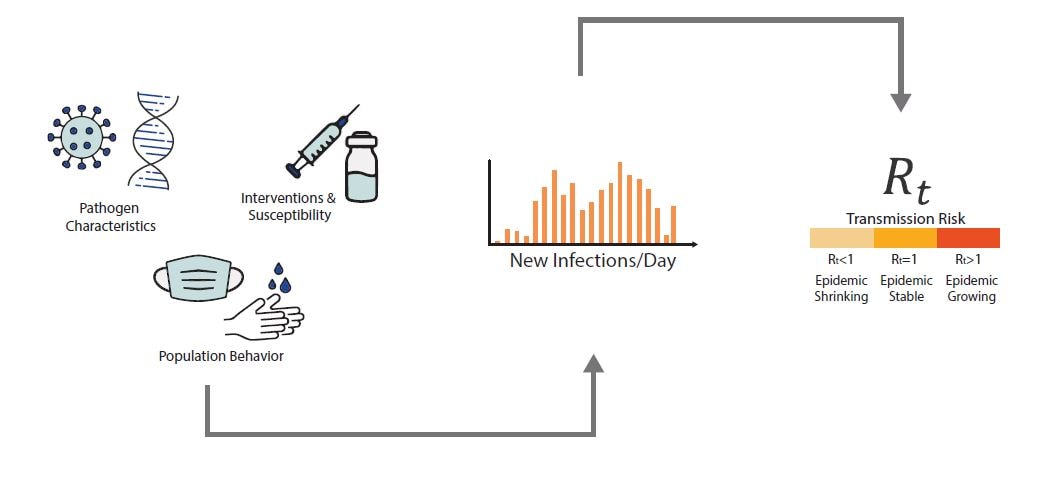
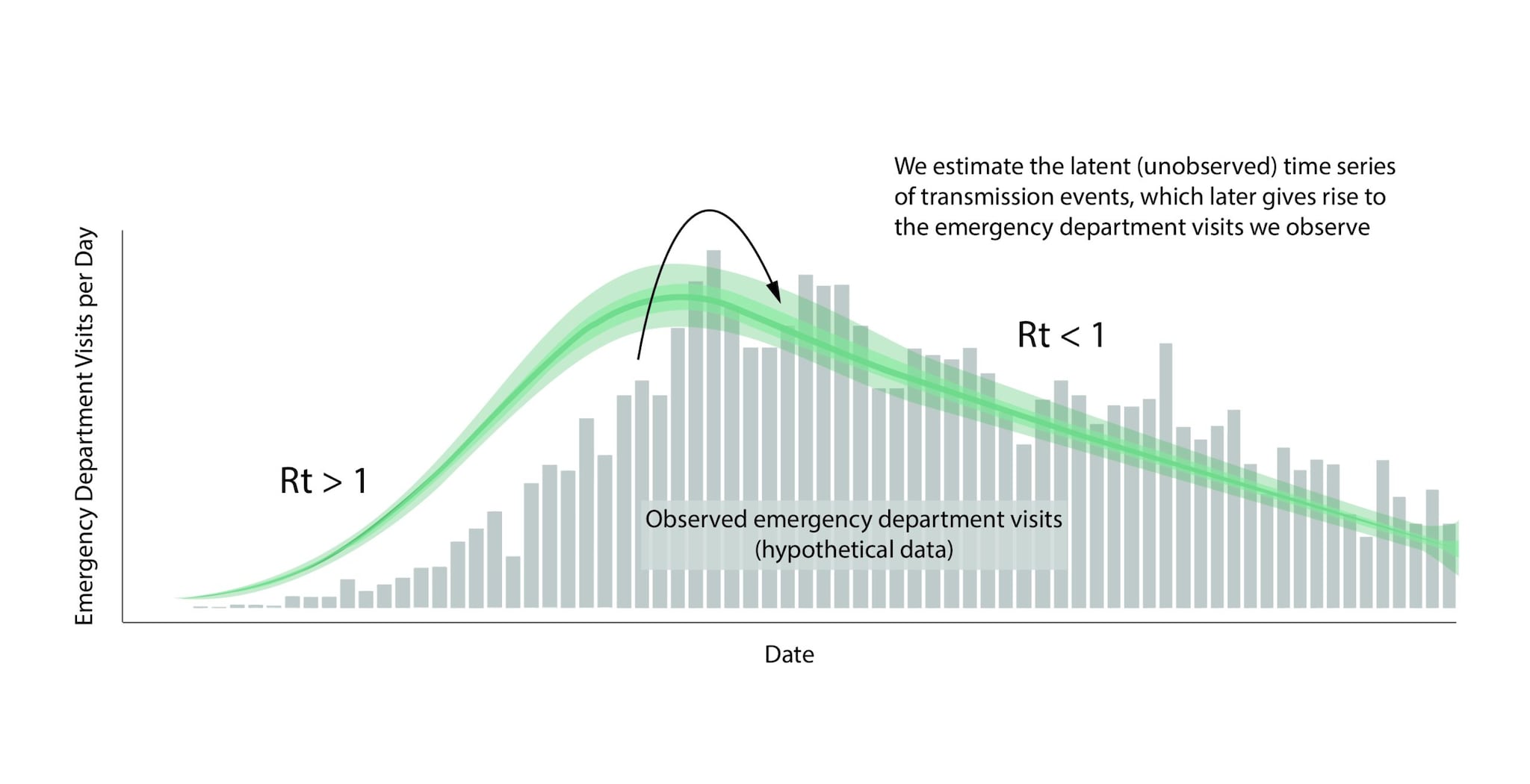
An epidemic's growth and decline are driven by underlying changes in transmission over time. Early in an epidemic of a novel disease, when everyone is susceptible, transmission rates are usually highest. Rates then decline as people change their behavior to avoid infection, or gain immunity through infection or immunization. The peak of an epidemic is a turning point that occurs when transmission falls below a critical threshold where, on average, each infected person no longer causes more than one new infection. Rt is defined as the average number of new infections caused by each infected person at time t, usually measured in days. Rt tells us whether, at that time, infections are increasing, decreasing, or staying relatively flat. We estimate Rt from data, and it accounts for current levels of population susceptibility, interventions, and behavior at the time the underlying infections occurred (Fig. 1). During an epidemic, Rt estimates provide information about the trend of the epidemic and can be used to forecast short-term changes in cases, hospitalizations, or deaths, and to assess the effectiveness of interventions designed to slow transmission.
For Public Health Professionals and Epidemiologists: Learn More About How We Estimate Rt
See Video featuring CDC Scientist Katie Gostic on how to calculate Rt
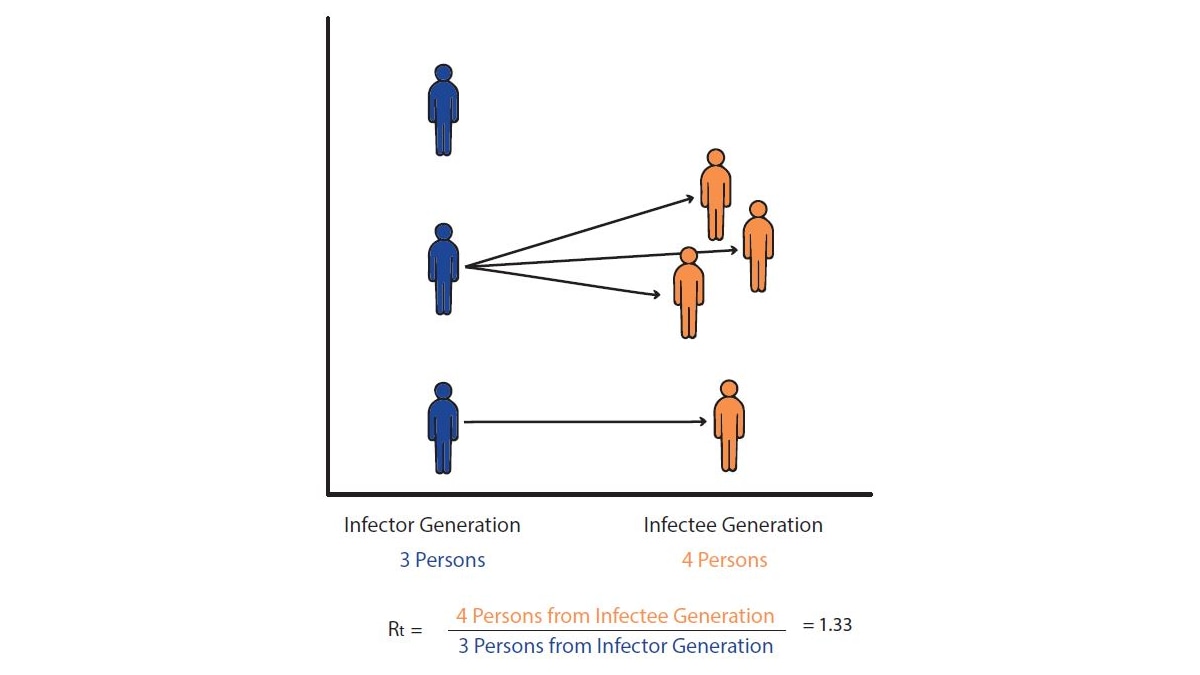
Let's imagine a world in which we can observe every disease transmission event exactly when it occurs. In this hypothetical world, we count the number of new infections that occurred on day t and divide by the number of infected persons who caused them, to give us Rt: the average number of new infections that each previously infected person caused (Fig. 2).
In reality, it's almost impossible to know exactly when transmission occurred or who infected whom in epidemiological data. While sometimes epidemiologists run focused studies designed to observe transmission events and transmission chains, these studies require intensive monitoring of a small group of participants and are the exception, not the rule. To get around these challenges, we estimate Rt using data that are relatively easy to obtain: daily counts of the number of new cases, emergency department visits, hospitalizations, or deaths. We input these data into a mathematical model designed to deal with three main challenges of data observation:
- We almost never know who infected whom
- There is a lag between the moment someone is infected (an unobservable event) and the date their infection could become observable and/or reported—e.g., as a symptomatic case, a positive test, an emergency department visit, or hospitalization.
- Not all infections will be observed. For example, not all cases will have symptoms and not all cases who have symptoms will be tested or will seek care at an emergency department.
Below is a walk-through of the basic logic, assumptions, and weaknesses of this model; those with more technical backgrounds may also wish to review the technical details of our approach.
To estimate Rt, we need to divide the total number of newly infected people on day t by the number of people who caused those infections (Fig. 2). But how can we do this if the data only contain counts of the total numbers of infections observed each day?
Note
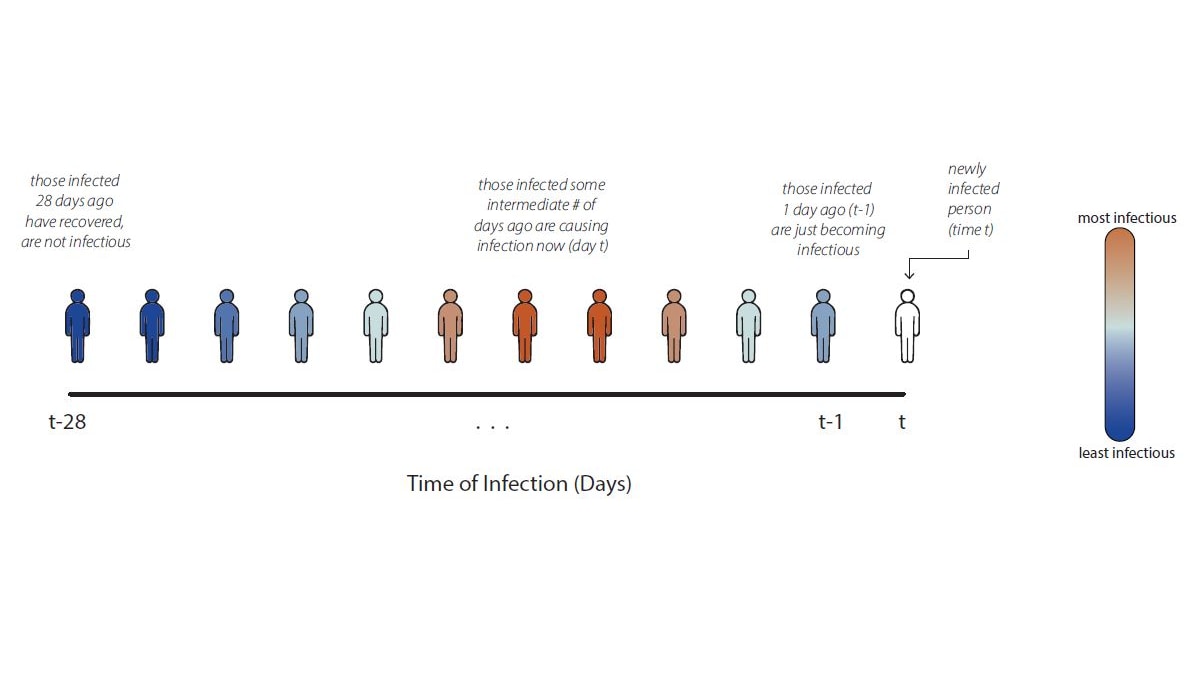
In count data, we can directly read the numerator of the Rt ratio—the number of newly infected people on day t—from the data. The denominator is more difficult to assess. Instead of trying to infer exactly who infected whom, we make assumptions grounded in infectious disease biology. For COVID-19, for example, we know that individuals infected yesterday are just becoming infectious as their viral loads increase. Meanwhile, individuals infected weeks ago have likely recovered and are no longer infectiousA. We can assume that individuals who were infected some intermediate number of days in the past are now causing the bulk of new infections (Fig. 3).
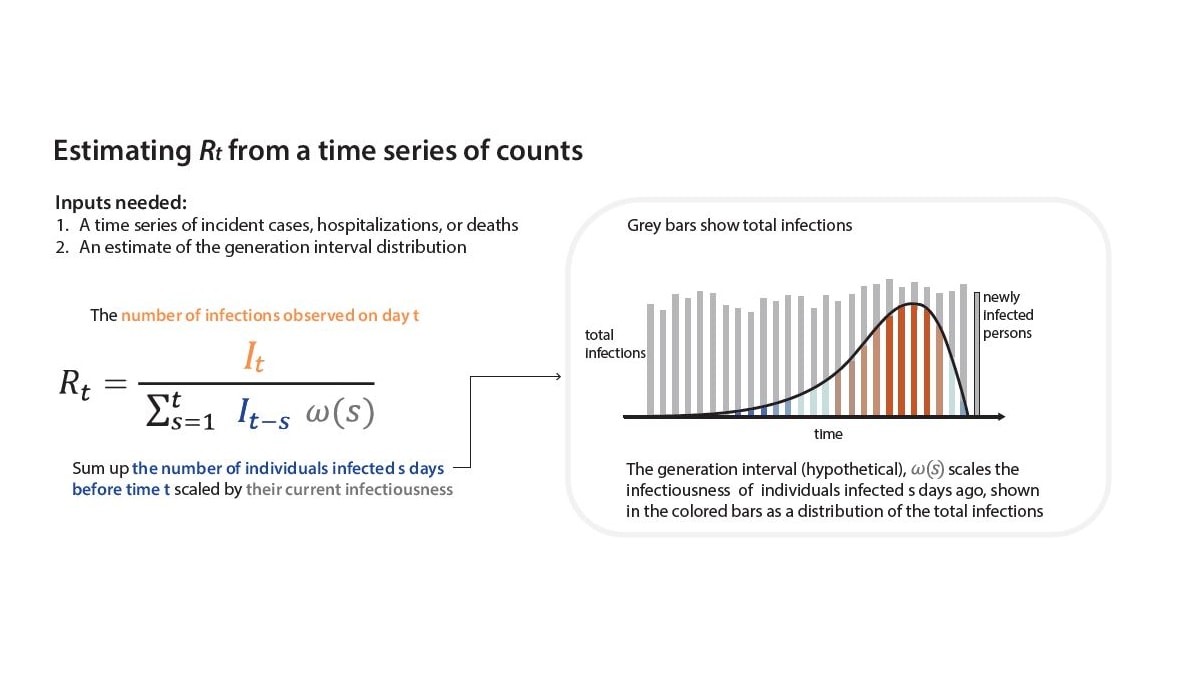
To estimate Rt, we must develop a model that turns the assumption "individuals infected some days in the past are the ones causing transmission now" into an equation. Our equation is a more complex version of the Rt ratio in Fig. 1, inferred using observable variables. To count the number of individuals in the infector generation on day t, we need to sum across all the individuals who became infected in the recent past—starting yesterday and going back weeks ago—weighted by their current infectiousness. For COVID-19, we assume that individuals infected between 1 and 7 days ago are most infectiousA, but individuals infected earlier or later may still cause infections. For details of our Rt equation, see Fig 4.
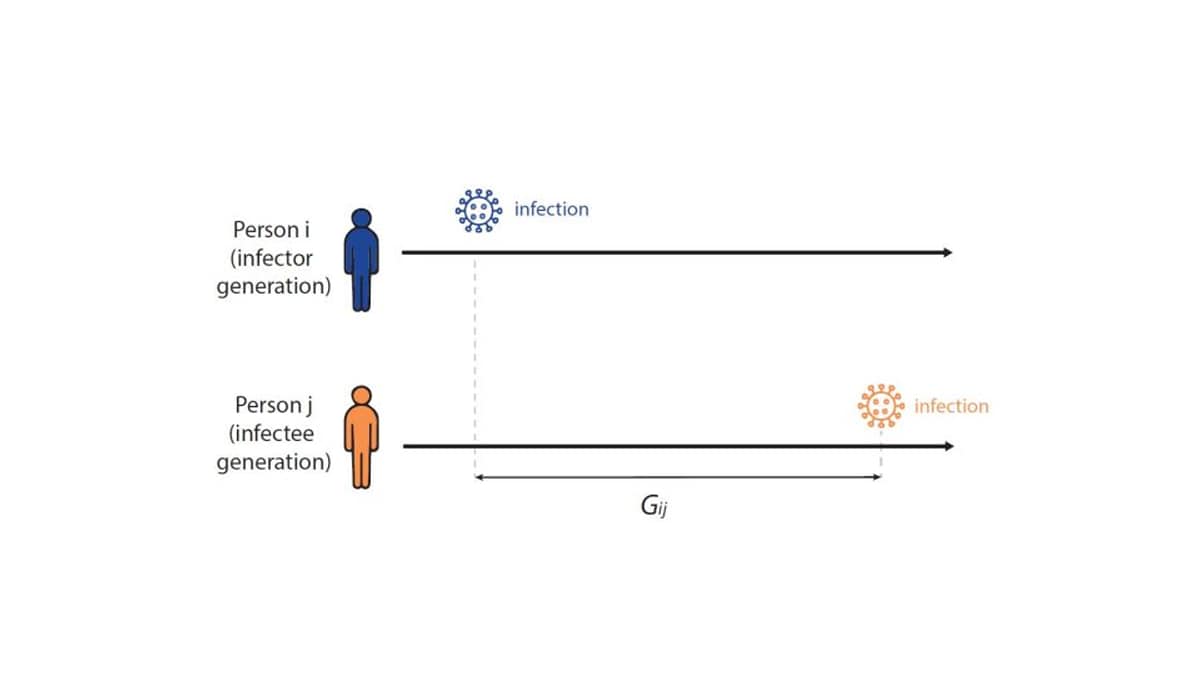
To formally estimate how long the expected wait between infections in a chain of transmission is—and to establish the infectiousness weighting function in Fig. 4 above—infectious disease models use a distribution called the generation interval (G), defined as the interval between the infection times of an infector-infectee pair (Fig 5). For example, if person i was infected on Monday, and if person i infects person j on Friday, then the Gij is four days. We know that the generation interval varies between transmission pairs, and so we want the distribution of times between infector-infectee pairs. We can estimate the generation interval distribution using data from household or contact tracing studies, in which the approximate timing of infections is observed, or by using the serial interval (the time between onset of symptoms of an infector-infectee pair) as a proxy.
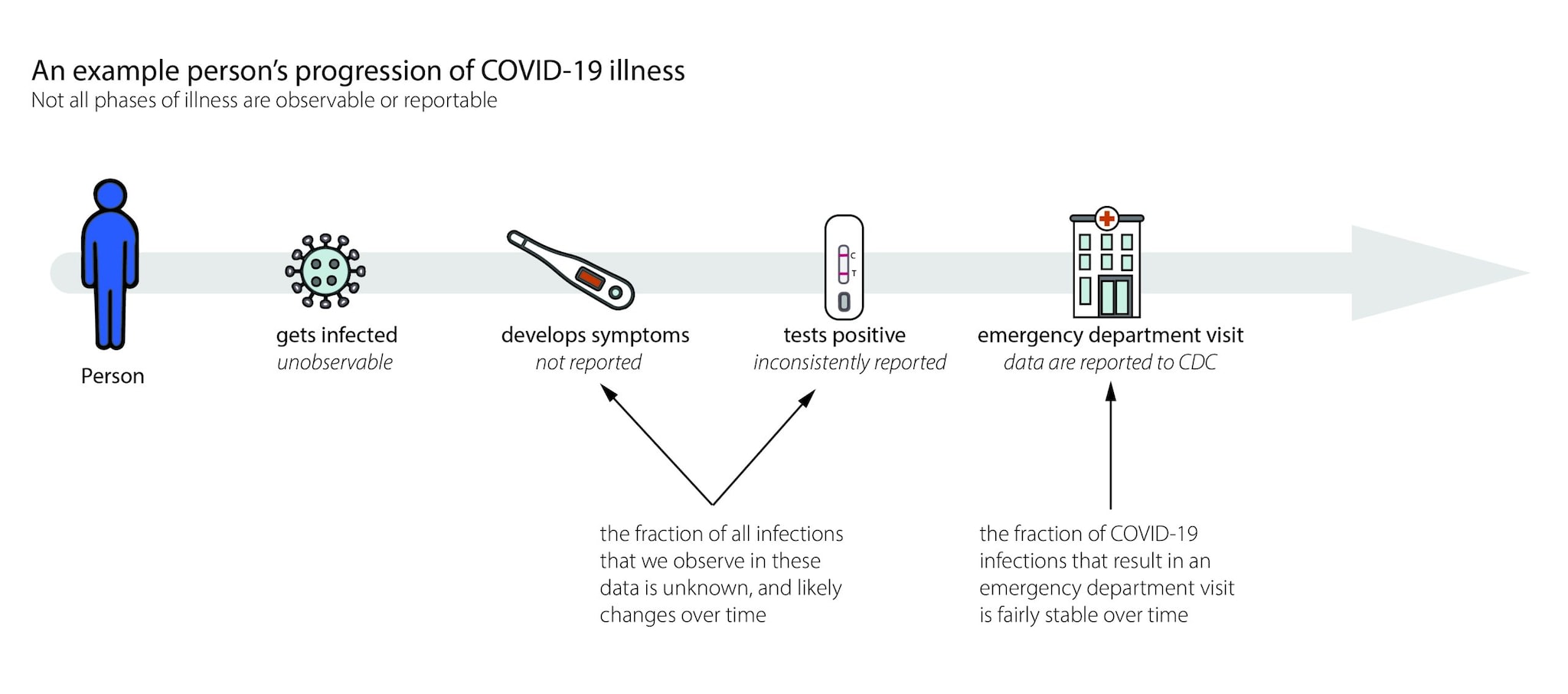
When we estimate Rt,we want to know how many new infections occur on day t. In the real world, we observe events like cases, emergency department visits, hospitalizations, or deaths with delays of days to weeks (Fig. 6). These delays are unavoidable and fall into two main categories:
- Biological delays, between the moment a person is first infected and the moment their infection could become observable and/or reportable as a confirmed case, emergency department visit, hospitalization, or death, and
- Reporting delays, between the time a person tests positive, visits an emergency department, is admitted to the hospital, or dies and the time that event is reported to the health department. Data from some events, like positive at-home tests, are often never reported and are therefore difficult to reliably or completely count (Fig. 6).
Caveats and complications:
- On the most recent dates, we have not yet observed all infections that have occurred, as some infected people have not yet developed symptoms or visited an emergency department. This is a challenge because people are usually most interested in recent trends, but recent data are incomplete.
- There are day-of-week effects in healthcare visits and reporting, where the data consistently show more reports on weekdays vs. weekends.
- Events (e.g., positive tests, emergency department visits, hospitalizations, deaths) are not always reported on the day that they occur. For example, sometimes test results take a few days to come back from the lab, diagnoses undergo review, or there are delays in transferring data.
To adjust for incomplete reporting on recent dates, CDC is implementing "nowcasting" approaches. Essentially, we can look back at past reporting patterns to estimate the fraction of total reports that were observed 1, 2,..., n days after the reported event. Then we can scale up accordingly to estimate the number of events that will eventually be reported on each day.
In our data, we only observe the fraction of infected individuals that visited the emergency department and were diagnosed with that infection (Fig. 7). Emergency department visit data are reported through the National Syndromic Surveillance Program (NSSP). Participation in the NSSP is voluntary, but many facilities that participate have automated reporting systems, and at the state level, total reported visit volumes are usually very stable over time. We work closely with partners at NSSP to monitor reported total visit volumes, and we manually review the reported data each week to ensure that reported Rt values reflect actual increases or decreases in transmission, not just increases or decreases in the number of reported emergency department visits. In our review of total reported emergency department visits, if we suspect there's an issue with the data—that data may be missing, that a facility has temporarily stopped reporting visits, or that a facility has recently been added to the system—we can clean the data. Data cleaning may include dropping one or two unusually low (or high) reported values, filtering which facilities are included in calculated totals, or—in the case of a more widespread reporting outage— choosing not to report an estimate for the affected jurisdiction until the problem is resolved.
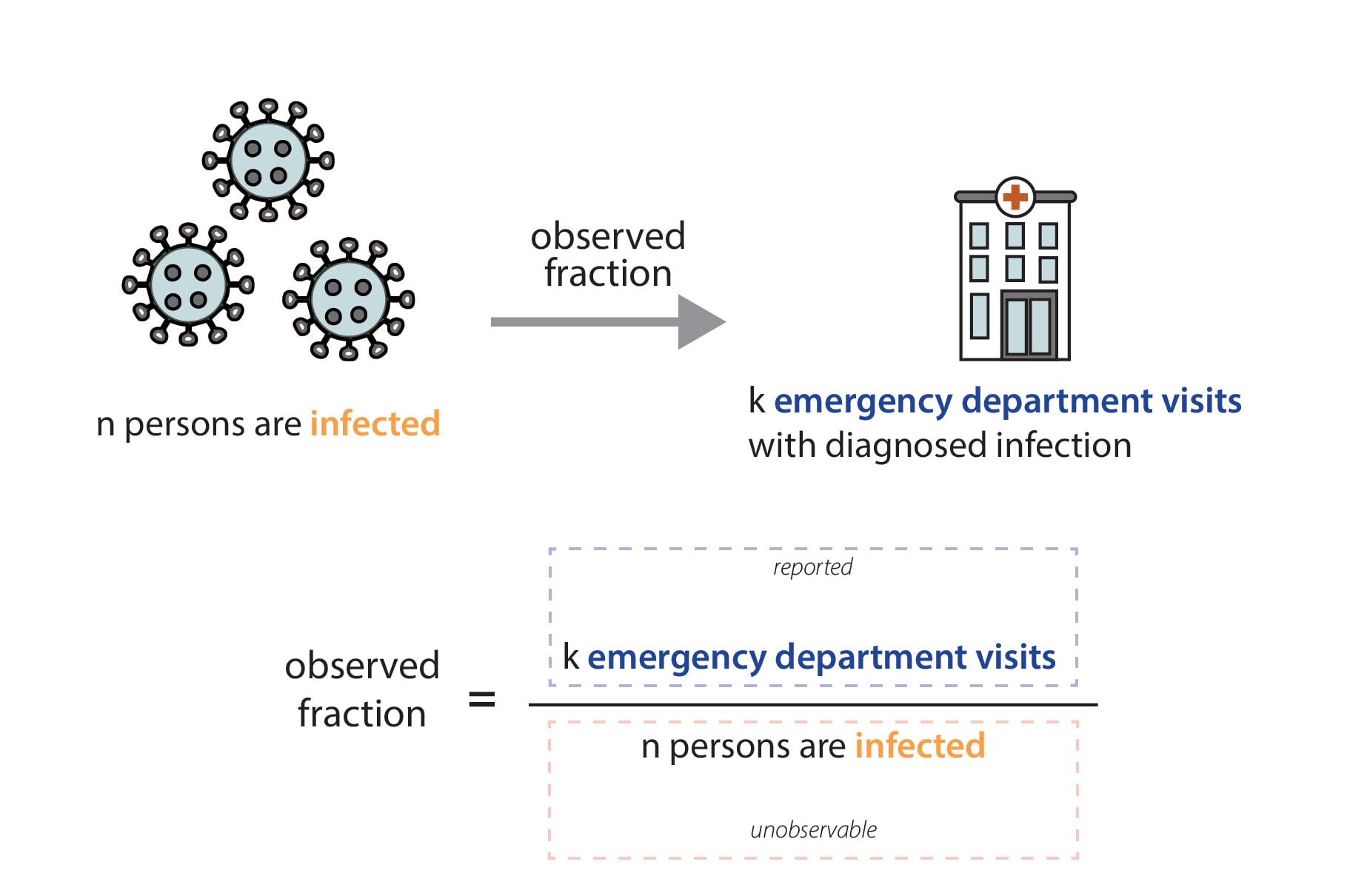
Mathematically, we expect our Rt estimates to be unbiased as long as the fraction of observed infections in emergency department visit data is not changing rapidly. That is because the observed fraction impacts both the numerator (the infectee generation) and denominator (the infector generation) of our Rt equation equivalently (Fig. 8). In reality, there is probably no epidemic dataset where there is no change at all in the fraction of observed infections over time. We have chosen to focus on emergency department visits because it is a stable signal, with widespread coverage across the United States, and because we think that focusing on the number of people seeking treatment at an emergency department allows us to observe one of the most stable fractions of infections of any available data source (Evaluating Data Types, 2020). There are some situations where the fraction of observed infections could change quickly enough to temporarily cause biases in the Rt estimates, such as the emergence of a more severe variant, lack of diagnostic tests, or a clinical or testing practice change within a healthcare setting. If such a situation occurs, we will flag the estimates we publish noting that there is some possibility of inaccuracy during the brief period before the fraction observed re-stabilizes.
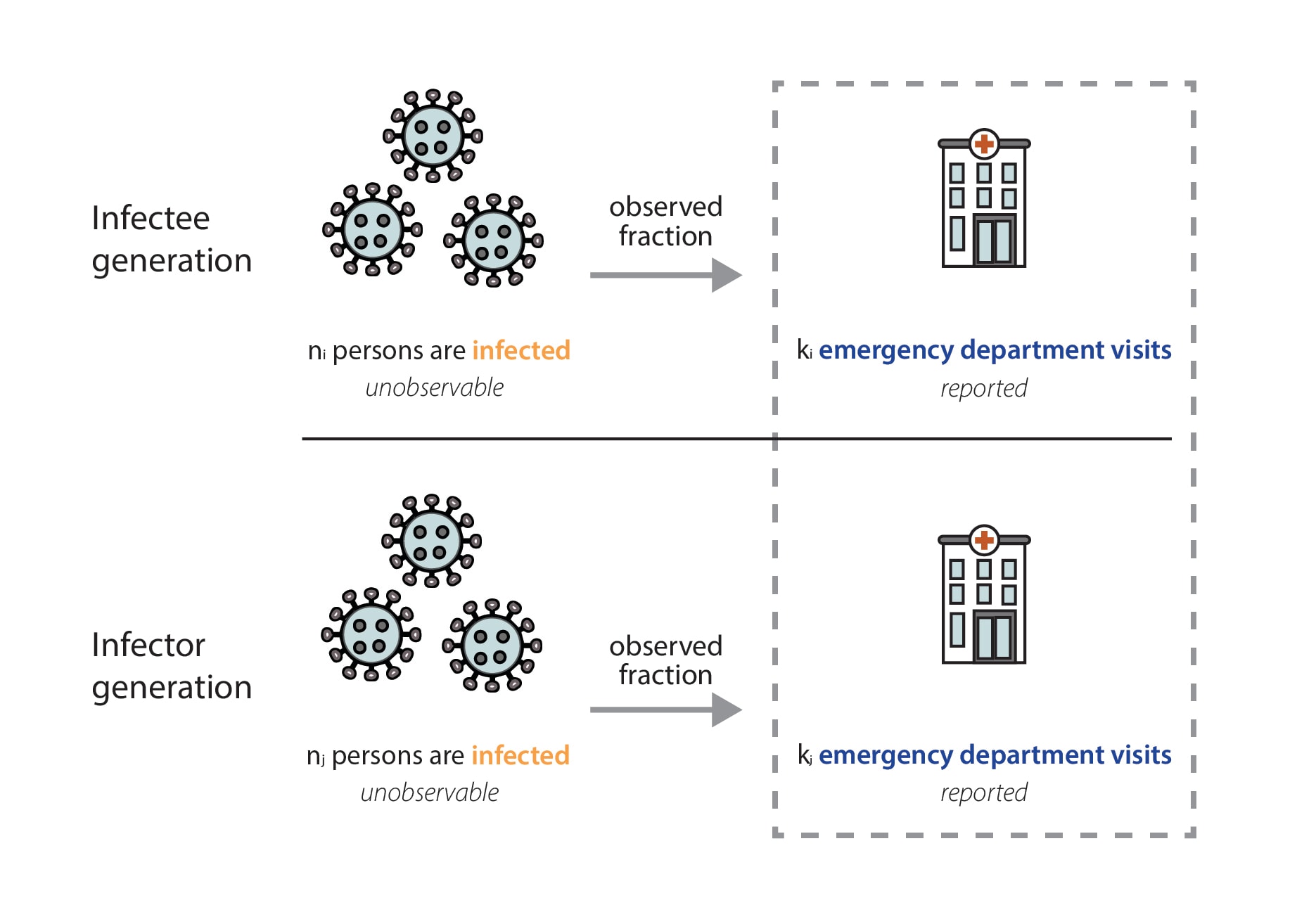
It is important to note that individuals visiting the emergency department for a respiratory illness could be systematically different from the general population; for example, they may be older, have other coexisting medical conditions, or limited access to other healthcare options (e.g., primary care or urgent care). However, these differences don't directly affect our estimates, because we are not measuring the number of new infections that specific individuals go on to cause. Instead, our estimates reflect the population average level of transmission that caused those individuals to become infected themselves.
In fact, though counterintuitive, mathematically, in an epidemic system without rapid changes in severity, infectiousness, or precautionary behavior, different age groups should experience roughly similar epidemic growth rates over time after an initial mixing period. While early in the COVID-19 pandemic these conditions were probably not met, at this point we believe these effects are minor. Although the total number of infections in each group will be different, the relative change should be the same. This means that estimates of Rt based on incident events from a subgroup (individuals who visit the emergency department, for example) of a population are unbiased as long as the fraction of observed infections in that subgroup stays roughly constant.
Rt Estimation Model |
||
| COVID-19 and Influenza | ||
| EpiNow2 | We use the epinow() function in the EpiNow2 R package to estimate Rt. The code we use to run the model is available on GitHub, and the package documentation explains the math behind the model. | |
Priors |
||
| COVID-19 and Influenza | ||
| We use the default prior values defined in the EpiNow2 package documentation, with the following exceptions: | ||
| Rt=0 (the initial Rt value) | Mean: 1, standard deviation (sd): 0.2 | |
| αsd (the standard deviation of the Gaussian process) | 0.0075 | |
| In very rare cases, we may adjust other model priors to improve sampler diagnostics and ensure convergence. Specifically, during the 2023-2024 respiratory virus season, we occasionally increased the mean of the prior on the Gaussian process length scale from 21 days to 30 days. This affected fewer than 10 model runs out of thousands (each run was specific to a jurisdiction, date, and disease). | ||
Disease-specific parameters |
||
| COVID-19 | ||
| Generation interval distribution | Lognormal distribution with mean 2.9 days and standard deviation 1.64 days (Park et al. 2003), discretized with maximum 12 days and minimum 1 day. (“generation_time” parameter in EpiNow2) | |
| Incubation period distribution | Modified Weibull distribution with mean 4.24 days (Park et al. 2003) | |
| Distribution of delays from symptom onset to emergency department (ED) visit | Lognormal distribution with mean 4.17 days and standard deviation 6.27 days, discretized with maximum 30 days and minimum 0 days (from internal data) | |
| Distribution of delays from infection to ED visit | Derived by combining the incubation period and delay from symptom onset to ED visit. (“delays” parameter in EpiNow2) | |
| Distribution of delays from date of ED visit to date of report | Jurisdiction-specific distribution, estimated from the average of observed reporting delays in the recent past (“truncation” parameter in Epinow2; used to adjust for incomplete reporting on recent dates) | |
| Influenza | ||
| Generation interval distribution | Gamma distribution with mean 3.52 days and standard deviation 2.10 days (Chan et al. 2024), discretized with maximum 9 days and minimum 1 day. (“generation_time” parameter in EpiNow2) | |
| Incubation period distribution | Gamma distribution with mean 1.55 days and standard deviation 0.66 days (Chan et al. 2024), discretized with a maximum of 5 days, and a minimum of 0 days. | |
| Distribution of delays from symptom onset to ED visit | Gamma distribution with mean of 3.52 days and standard deviation of 2.91 days, discretized with maximum 15 days and minimum 0 days (from internal data) | |
| Distribution of delays from infection to ED visit | Derived by combining the incubation period and delay from symptom onset to ED visit. (“delays” parameter in EpiNow2) | |
| Distribution of delays from date of ED visit to date of report | Jurisdiction-specific distribution, estimated from the average of observed reporting delays in the recent past (“truncation” parameter in Epinow2; used to adjust for incomplete reporting on recent dates) | |
In some epidemic modeling analyses, we get to check our answers. For example, if we generate a short-term epidemic forecast, we can wait a few weeks, and then check our predictions against what really happened. But we're never able to observe Rt directly, and so we don't have a gold standard source of truth to check our models against. As a result, we use a few different methods to check that our estimates are reliable.
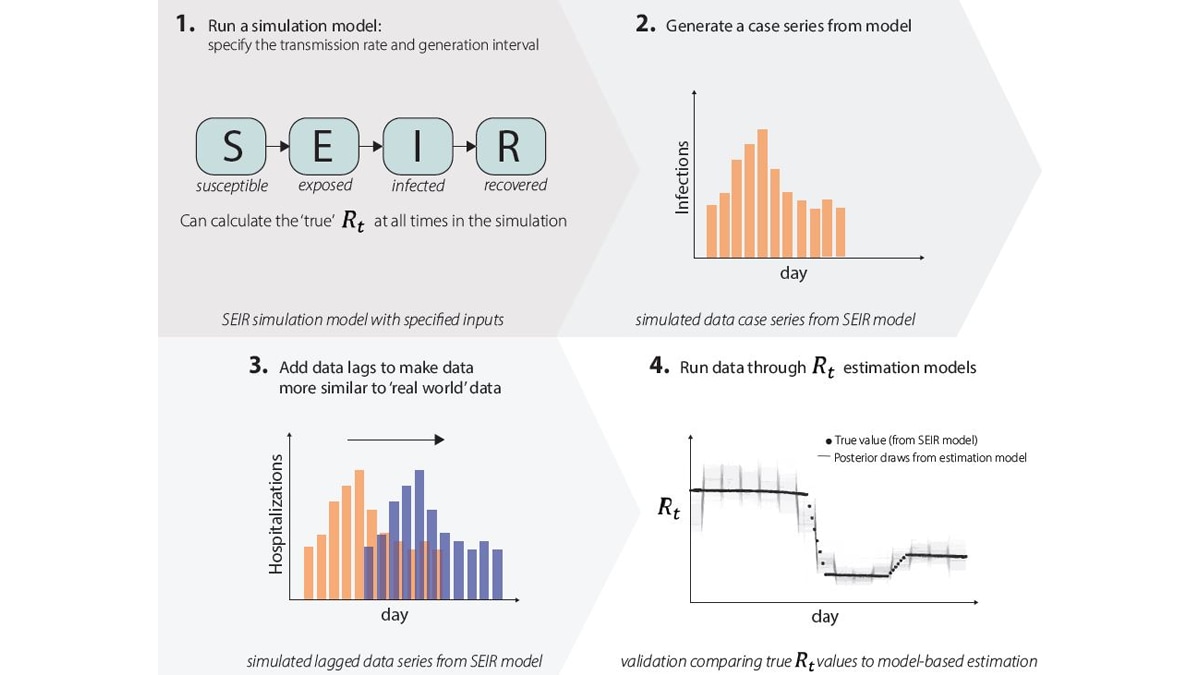
1. We run simulation studies. We run an epidemic simulation using a dynamic mathematical model with four compartments: susceptible (S), exposed (E), infected (I), and recovered (R) (Fig. 9.1) where we can calculate the 'true' Rt value at all times. The simulation produces an epidemic time series with counts of the number of new infections per day (Fig. 9.2), and we add lags to these data to make them more similar to the case, hospitalization, or death data that we observe in the real world (Fig. 9.3). We can run these simulated data through our Rt estimation models just like real data, only in this case we know exactly what the answer (Rt) should be, as we specified it when simulating the data. We then compare results to the correct answers (Fig. 9.4). If our models do not accurately estimate Rt, we know we need to make changes until the model accurately estimates Rt.
2. We check that our real-time estimates are consistent with observed trends. We compare estimates with each other to ensure they are reasonably consistent over time as new data become available.
3. We perform common-sense checks. If the data show that the epidemic is growing rapidly, then we should see Rt estimates, including confidence intervals, above one for the corresponding time period, after adjusting for lags.
4. We evaluate nowcasts from our models. We validate that the models consistently estimate final reports accurately, using the partial information available at the time.
Key Takeaways
Key Points
Rt is a transmission metric that estimates the ratio of infected to infectors in an epidemic at a particular point in time. Rt estimates help inform situational awareness, giving clues as to how quickly an epidemic is likely to increase or decrease in the near future. To be useful for decision making, Rt estimates need to be accurate, accounting for time lags because transmission events causing cases now occurred days to weeks ago (Fig. 10).
Especially in a novel outbreak, it is essential to know whether the epidemic has started, if we are nearing a peak, and/or if transmission has begun to decline. Rt allows policymakers and public health decision-makers to assess the impact of interventions because it estimates how transmission rates have changed over time, and to assess the intensity of spread because it directly reflects growth in infections.
What’s next?
We are estimating Rt for respiratory viruses in collaboration with the National Center for Immunization and Respiratory Diseases. In the longer term, we plan to build a well-tested analytic infrastructure that we could use to estimate Rt for a novel pathogen in a future infectious disease epidemic. Even for something like Rt, where the quantity we're trying to estimate is conceptually relatively simple, it takes incredible care and sophisticated modeling tools to adjust data as they are observed and obtain accurate estimates quickly. We are also exploring new models that will allow us to combine wastewater data with other signals when we estimate Rt. It is incredibly difficult to build these kinds of analytic pipelines on the fly. Investing the time now to build good infrastructure and think through the problems we can anticipate will leave us better prepared for the next infectious disease epidemic.
- Day 1-7 covers the central 95% of the generation interval distribution for Omicron from Park et al., 2023; day 1-5 covers the central 80% of the generation interval distribution. See Figure 5 for a definition of the generation interval.
