Purpose
This step-by-step guide—complete with code examples—will walk public health professionals through the process for SQL queries on the BioSense Platform using Posit Workbench.

A step-by-step guide
You can do all your SQL queries on the BioSense Platform by using Posit Workbench. (Posit is the tool formerly known as RStudio.) Here are some easy steps to follow and code examples for querying data on the BioSense Platform's DataMart.
First, sign in to your Posit Workbench account using your Access & Management Center (AMC) username and password (Figure 1).

Next, install and load the packages that you'll need, which are called libraries. These tell Posit Workbench how to connect to the Analytic DataMart. To do this, copy and paste the next two lines into your code.
library(DBI)
library(odbc)
To run pieces of code, you can highlight the code in your R script and use Ctrl + Enter or use the Run button.
Now you can connect to the Analytic DataMart. This step uses the username and password that you're already using to connect to Posit Workbench, so there's no need to re-enter them. You can even store this connection as a variable to reuse.
Copy and paste the line below to store a database connection directly to the BioSense Platform database on the Analytic DataMart.
datamart <- dbConnect(odbc::odbc(), dsn = "BioSense_Platform")
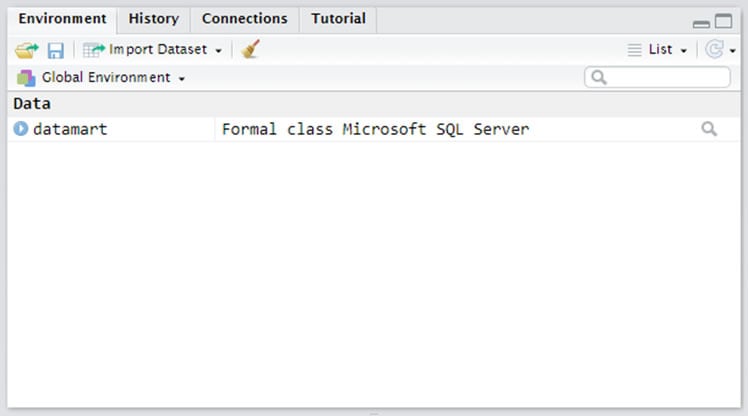
You can confirm that you're connected to the DataMart by looking at the Environment pane on the right side of your screen (Figure 2).
The last step is to write your query and send it to the database. To do this, copy and paste the code below to R:
table <- dbGetQuery(datamart, "SELECT * FROM Except_Reasons"))
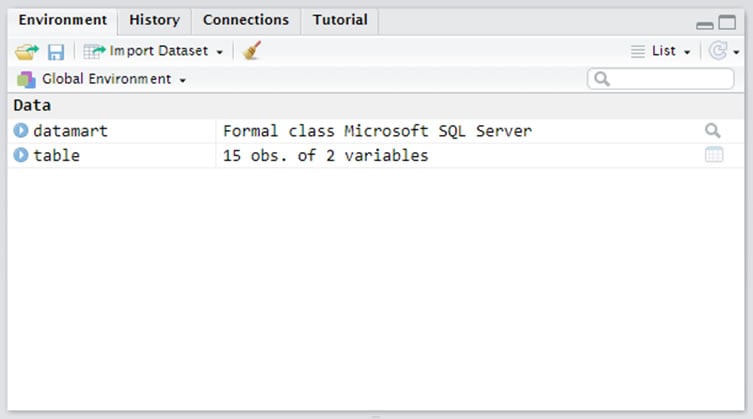
This example pulls information from the Except Reasons table, but you can insert any query you'd like inside the quotation marks. Once the query is complete, the table of query results will appear in your Environment pane (Figure 3).
And that's it! The code shown below is enough to get you started reviewing your data in Posit Workbench. After you copy and paste this code into Posit Workbench, delete and retype all double-quotes (").
library(DBI)
library(odbc)
datamart <- dbConnect(odbc::odbc(), dsn = "BioSense_Platform")
table <- dbGetQuery(datamart, "SELECT * FROM Except_Reasons")
write.csv(table, file ="TableName.csv", row.names = FALSE)
The write.csv command will write the results to a .csv file that can be exported and opened as an Excel file.
